Amyotrophic Lateral Sclerosis (ALS) and Frontotemporal Dementia (FTD) Panel
Summary and Pricing 
Test Method
Exome Sequencing with CNV Detection| Test Code | Test Copy Genes | Panel CPT Code | Gene CPT Codes Copy CPT Code | Base Price | |
|---|---|---|---|---|---|
| 10359 | Genes x (38) | 81479 | 81179(x1), 81404(x1), 81405(x2), 81406(x7), 81407(x1), 81479(x64) | $990 | Order Options and Pricing |
Pricing Comments
C9orf72 hexanucleotide repeat expansion will be performed first as this is the most common cause of ALS and FTD. If C9orf72 testing is negative then the sequencing panel and ATXN2 repeat expansion testing will be performed. Results for the C9orf72, ATXN2, and sequencing panel test will be reported separately. Concurrent testing is available upon request. C9ORF72 and ATXN2 repeat expansions are available separately if needed.
We are happy to accommodate requests for testing single genes in this panel or a subset of these genes. The price will remain the list price. If desired, free reflex testing to remaining genes on panel is available. Alternatively, a single gene or subset of genes can also be ordered via our Custom Panel tool.
An additional 25% charge will be applied to STAT orders. STAT orders are prioritized throughout the testing process.
Click here for costs to reflex to whole PGxome (if original test is on PGxome Sequencing platform).
Click here for costs to reflex to whole PGnome (if original test is on PGnome Sequencing platform).
Turnaround Time
C9orf72 assays completed within 2 weeks on average. Reflexive panels are completed within an additional 4 weeks on average.
Please note: Once the testing process begins, an Estimated Report Date (ERD) range will be displayed in the portal. This is the most accurate prediction of when your report will be complete and may differ from the average TAT published on our website. About 85% of our tests will be reported within or before the ERD range. We will notify you of significant delays or holds which will impact the ERD. Learn more about turnaround times here.
Targeted Testing
For ordering sequencing of targeted known variants, go to our Targeted Variants page.
Clinical Features and Genetics 
Clinical Features
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease characterized by motor neuron impairment in the cortex, brain stem, and spinal cord (Hardiman et al. 2017. PubMed ID: 28980624). The dysfunction and loss of these neurons results in rapid progressive muscle weakness, atrophy and ultimately paralysis of limb, bulbar and respiratory muscles. About 50% of patients also develop cognitive and behavioral impairment and another 13% develop frontotemporal dementia (van Es et al. 2017. PubMed ID: 28552366). The mean age of symptom onset is 55 years of age for familial cases and 65 years of age for sporadic cases; most cases begin between 40 and 70 years of age. The annual incidence of ALS is 2-3 per 100,000 (van Es et al. 2017. PubMed ID: 28552366; Brown and Al-Chalabi. 2017. PubMed ID: 28700839; Siddique and Siddique. 2019. PubMed ID: 20301623).
The most common ALS symptoms include twitching and cramping of muscles of the hands and feet, loss of motor control in the hands and arms, weakness and fatigue, tripping and falling. Symptoms usually begin with asymmetric involvement of the muscles. As the disease progresses, symptoms may include difficulty in talking, breathing and swallowing, shortness of breath, and paralysis.
Frontotemporal dementia (FTD) is a clinically heterogeneous syndrome due to the progressive degeneration and atrophy of various regions of the frontal and temporal lobes of the brain. Symptoms are variable with average age of onset in the sixth decade of life; although earlier and later onsets have been documented (Neary et al. 1998. PubMed ID: 9855500; Snowden et al. 2002. PubMed ID: 11823324; Bruni et al. 2007. PubMed ID: 17620546). The annual incidence of FTD is 3-4 per 100,000 (Onyike and Diehl-Schmid. 2013. PubMed ID: 23611343).
Two major forms of FTD, the behavioral-variant (FTD-bv) and the primary progressive aphasia (PPA), are recognized based on the site of onset of degeneration and the associated symptoms. FTD-bv comprises about half of FTD cases with the degenerative process beginning in the frontal lobes and resulting in personality changes and deterioration of social conducts. Most common behavioral changes are: disinhibition, apathy, deterioration of executive function, obsessive thoughts, compulsive behavior, and neglect of personal hygiene (Rascovsky et al. 2011. PubMed ID: 21810890). PPA is a language disorder that is further divided into two sub-forms: progressive nonfluent PPA and semantic variant PPA. Nonfluent PPA is characterized by difficulty in verbal communications, word retrieval, and speech distortion. Reading, writing and spelling are also affected; while memory is relatively preserved. Semantic variant PPA is characterized by the progressive impairment of word comprehension, object and face recognition, and loss of semantic memory. Reading and writing skills are relatively preserved (Gustafson. 1993. PubMed ID: 8401782).
Genetics
About 10% of ALS cases and 30-50% of FTD cases are inherited. In most of these families, ALS and/or FTD is inherited in an autosomal dominant manner with age-dependent penetrance. In rare families, the disease is transmitted in an autosomal recessive or dominant X-linked pattern via the UBQLN2 gene. About 90% of patients with ALS are sporadic cases (SALS) with no known affected relatives. The C9orf72, SOD1, FUS, and TARDBP gene account for 35%, 15%, 3% and 4% of European familial ALS (FALS) cases. In Asian FALS patients, pathogenic variants in the C9orf72, SOD1, FUS, TARDBP genes account for 2%, 30%, 6% and 1.5% cases (Zou et al. 2017. PubMed ID: 28057713).
An expansion of a GGGGCC hexanucleotide repeat in a non-coding region of the C9orf72 gene is the most common inherited cause of ALS and FTD (Renton et al. 2011. PubMed ID: 21944779; DeJesus-Hernandez et al. 2011. PubMed ID: 21944778). Alleles with less than 25 repeats are usually considered normal (Majounie et al. 2012. PubMed ID: 22406228; van der Zee et al. 2013. PubMed ID: 23111906). Alleles with more than 30 repeats are considered pathogenic (Renton et al. 2011. PubMed ID: 21944779). This expanded repeat accounts for up to 40% of FALS cases and 7% of SALS cases (Byrne et al. 2012. PubMed ID: 22305801). The GGGGCC hexanucleotide repeat expansion was reported in patients with ALS, ALS-FTD or FTD (Siddique and Siddique. 2019. PubMed ID: 20301623).
ATXN2 expansions of the exon 1 poly-glutamine track ranging from 29-32 CAG repeats have also been associated with an increased risk for development of ALS (Van Damme et al. 2011. PubMed ID: 21562247). While expansions >32 repeats are typically associated with Spinocerebellar Ataxia 2 (SCA2), patients with ALS and with >32 repeats have been reported (Sproviero et al. 2017. PubMed ID: 28017481; Ross et al. 2011. PubMed ID: 21610160). For example, ALS was reported in a family with one affected individual being homozygous for 33 repeats and a second affected individual being compound heterozygous for 31 and 33 repeats (Van Damme et al. 2011. PubMed ID: 21562247). Penetrance is incomplete for individuals with 29-32 repeats.
FTDs are inherited in 30-50% of cases in an autosomal dominant manner with pathogenic variants in C9orf72, MAPT and GRN accounting for about 30%, 15% and 25% of cases respectively. An expansion of a GGGGCC hexanucleotide repeat in an intron 1 non-coding region of the C9orf72 gene is the most common inherited cause of FTD.
See individual gene summaries for more information about molecular biology of gene products and spectra of pathogenic variants.
Clinical Sensitivity - Sequencing with CNV PGxome
This NGS panel will detect pathogenic variants in at least 68% of patients with FALS and 11% of SALS. Pathogenic variants in the C9orf72 (40%), SOD1 (1-50%), FUS (1-5%), and TARDBP (1-5%) genes account for the majority of FALS cases. In SALS C9orf72 and SOD1 account for 7% and 1-2% of cases respectively (van Es et al. 2017. PubMed ID: 28552366; Robberecht and Philips. 2013. PubMed ID: 23463272). Pathogenic variants in the ANG, SQSTM1, PFN1, UBQLN2, OPTN, VCP, and VAPB genes account for less <1% of FALS cases. Clinical sensitivity for the C21orf2, CHCHD10, CHMP2B, DCTN1, ERBB4, FIG4, HNRNPA1, HNRNPA2B1, KIF5A, MATR3, MOBP, NEFH, NEK1, SCFD1, SETX, TAF15, TBK1, TUBA4A, and UNC13A genes is currently unknown due to the limited number of cases reported to date, but is expected to be low (Robberecht and Philips. 2013. PubMed ID: 23463272; Brown and Al-Chalabi. 2017. PubMed ID: 28700839).
In familial FTD, pathogenic variants are identified in up to 65% of cases with C9orf72, MAPT, and GRN accounting for about three-fourths of these cases (Goldman and Van Deerlin et al. 2018. PubMed ID: 29971646). The majority of genes in this panel have no or very few large deletions/duplications.
Testing Strategy
C9orf72 hexanucleotide repeat expansion will be performed first as this is the most common cause of ALS and FTD. If C9orf72 testing is negative then the sequencing panel and ATXN2 repeat expansion testing will be performed. Results for the C9orf72, ATXN2, and sequencing panel test will be reported separately. Concurrent testing is available upon request.
Both the C9orf72 and ATXN2 repeat expansion tests each utilize four unique gene specific assays: (1) a repeat primed PCR assay with the locus specific primer 5’ (upstream) of the repeat region (2) a repeat primed PCR assay with the locus specific primer 3’ (downstream) of the repeat region and two unique fluorescent fragment length assays. C9orf72 and ATXN2 are not included in the sequencing panel.
The sequencing panel typically provides 99.1% coverage of all coding exons of the genes listed plus 10 bases of flanking noncoding DNA in all available transcripts along with other non-coding regions in which pathogenic variants have been identified at PreventionGenetics or reported elsewhere. We define coverage as ≥20X NGS reads or Sanger sequencing. PGnome panels typically provide slightly increased coverage over the PGxome equivalent. PGnome sequencing panels have the added benefit of additional analysis and reporting of deep intronic regions (where applicable).
Dependent on the sequencing backbone selected for this testing, discounted reflex testing to any other similar backbone-based test is available (i.e., PGxome panel to whole PGxome; PGnome panel to whole PGnome).
Indications for Test
Patients with ALS typically present with upper and lower motor neuron impairment with other diagnoses excluded via imaging and neurophysiological examinations. Diagnosis can be difficult as sites of symptom onset and disease progression can be variable. Other screening tools used for ALS include the ALS-Brief Cognitive Assessement (ALS-BCA), the ALS Cognitive Behavioral Scressn (ALS-CBS) and the Edinburgh Cognitive and Behavioral ALS Screen (ECAS) (van Es et al. 2017. PubMed ID: 28552366).
The International Behavioral Variant FTD Criteria Consortium (FTDC) defines diagnosis for bvFTD as patients that present with three of the six clinical features: disinhibition, apathy, loss of sympathy, compulsive behaviors, hyperorality, and/or dysexecutive neuropsychologic profile. Diagnosis for PPA includes language deficits as the primary cause of impaired living and aphasia being most prominent at symptom onset.
Patients with ALS typically present with upper and lower motor neuron impairment with other diagnoses excluded via imaging and neurophysiological examinations. Diagnosis can be difficult as sites of symptom onset and disease progression can be variable. Other screening tools used for ALS include the ALS-Brief Cognitive Assessement (ALS-BCA), the ALS Cognitive Behavioral Scressn (ALS-CBS) and the Edinburgh Cognitive and Behavioral ALS Screen (ECAS) (van Es et al. 2017. PubMed ID: 28552366).
The International Behavioral Variant FTD Criteria Consortium (FTDC) defines diagnosis for bvFTD as patients that present with three of the six clinical features: disinhibition, apathy, loss of sympathy, compulsive behaviors, hyperorality, and/or dysexecutive neuropsychologic profile. Diagnosis for PPA includes language deficits as the primary cause of impaired living and aphasia being most prominent at symptom onset.
Genes
| Official Gene Symbol | OMIM ID |
|---|---|
| ALS2 | 606352 |
| ANXA11 | 602572 |
| APOE | 107741 |
| APP | 104760 |
| ATXN2 | 601517 |
| C9orf72 | 614260 |
| CHCHD10 | 615903 |
| CHMP2B | 609512 |
| DCTN1 | 601143 |
| DNAJC7 | 601964 |
| FUS | 137070 |
| GLE1 | 603371 |
| GRN | 138945 |
| ITM2B | 603904 |
| KIF5A | 602821 |
| MAPT | 157140 |
| MATR3 | 164015 |
| NEK1 | 604588 |
| OPTN | 602432 |
| PFN1 | 176610 |
| PSEN1 | 104311 |
| PSEN2 | 600759 |
| SETX | 608465 |
| SIGMAR1 | 601978 |
| SNCA | 163890 |
| SOD1 | 147450 |
| SPG11 | 610844 |
| SPTLC1 | 605712 |
| SPTLC2 | 605713 |
| SQSTM1 | 601530 |
| TARDBP | 605078 |
| TBK1 | 604834 |
| TREM2 | 605086 |
| TUBA4A | 191110 |
| UBQLN2 | 300264 |
| UNC13A | 609894 |
| VAPB | 605704 |
| VCP | 601023 |
| Inheritance | Abbreviation |
|---|---|
| Autosomal Dominant | AD |
| Autosomal Recessive | AR |
| X-Linked | XL |
| Mitochondrial | MT |
Diseases
Related Tests
Citations 
- Brown and Al-Chalabi. 2017. PubMed ID: 28700839
- Bruni et al. 2007. PubMed ID: 17620546
- Byrne et al. 2012. PubMed ID: 22305801
- DeJesus-Hernandez et al. 2011. PubMed ID: 21944778
- Goldman and Van Deerlin et al. 2018. PubMed ID: 29971646
- Gustafson. 1993. PubMed ID: 8401782
- Hardiman et al. 2017. PubMed ID: 28980624
- Majounie et al. 2012. PubMed ID: 22406228
- Neary et al. 1998. PubMed ID: 9855500
- Onyike and Diehl-Schmid. 2013. PubMed ID: 23611343
- Rascovsky et al. 2011. PubMed ID: 21810890
- Renton et al. 2011. PubMed ID: 21944779
- Robberecht and Philips. 2013. PubMed ID: 23463272
- Ross et al. 2011. PubMed ID: 21610160
- Siddique and Siddique. 2019. PubMed ID: 20301623
- Snowden et al. 2002. PubMed ID: 11823324
- Sproviero et al. 2017. PubMed ID: 28017481
- Van Damme et al. 2011. PubMed ID: 21562247
- van der Zee et al. 2013. PubMed ID: 23111906
- van Es et al. 2017. PubMed ID: 28552366
- Zou et al. 2017. PubMed ID: 28057713
Ordering/Specimens 
Ordering Options
We offer several options when ordering sequencing tests. For more information on these options, see our Ordering Instructions page. To view available options, click on the Order Options button within the test description.
myPrevent - Online Ordering
- The test can be added to your online orders in the Summary and Pricing section.
- Once the test has been added log in to myPrevent to fill out an online requisition form.
- PGnome sequencing panels can be ordered via the myPrevent portal only at this time.
Requisition Form
- A completed requisition form must accompany all specimens.
- Billing information along with specimen and shipping instructions are within the requisition form.
- All testing must be ordered by a qualified healthcare provider.
For Requisition Forms, visit our Forms page
If ordering a Duo or Trio test, the proband and all comparator samples are required to initiate testing. If we do not receive all required samples for the test ordered within 21 days, we will convert the order to the most effective testing strategy with the samples available. Prior authorization and/or billing in place may be impacted by a change in test code.
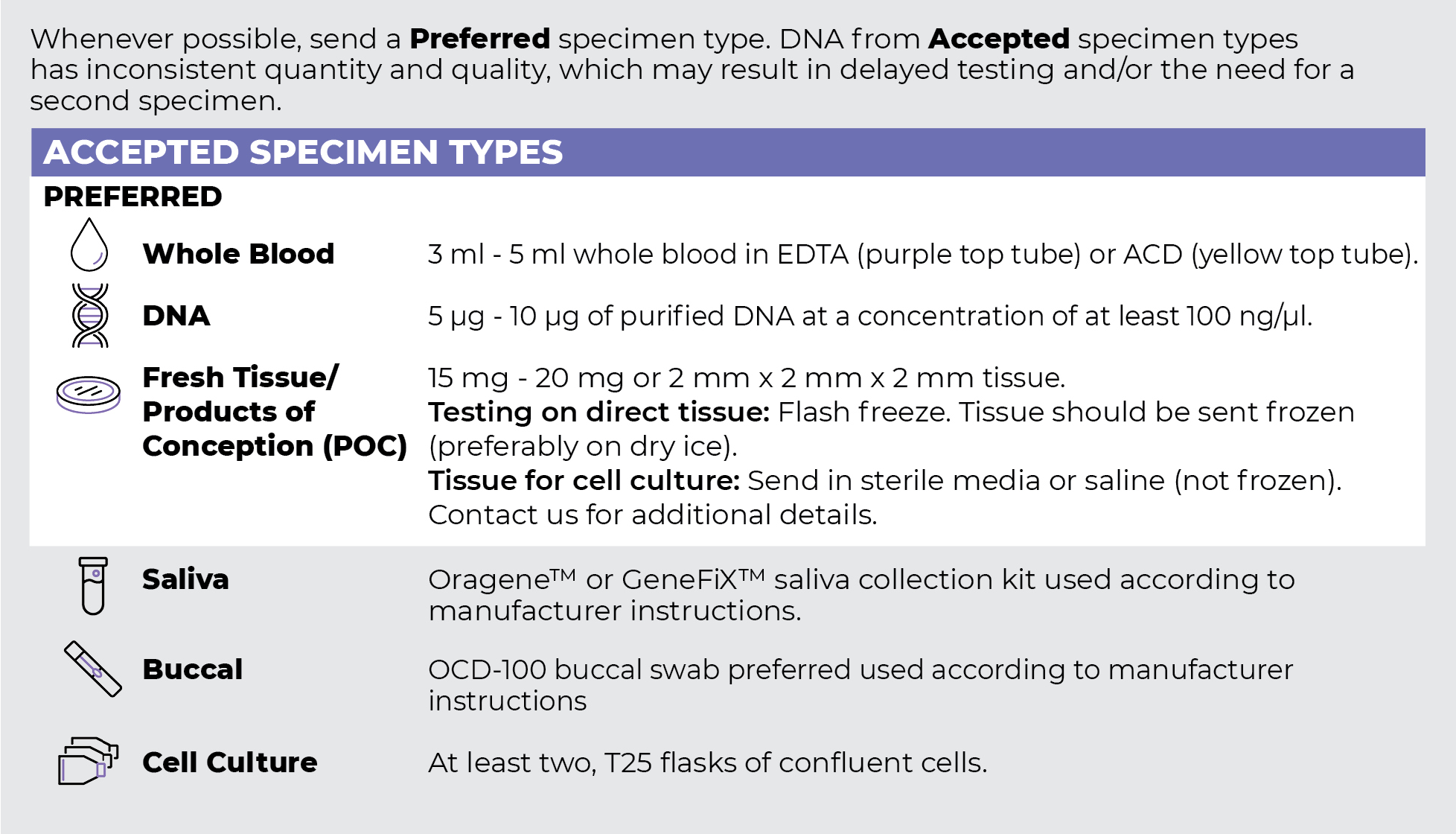
Specimen Types
Specimen Requirements and Shipping Details
PGxome (Exome) Sequencing Panel
PGnome (Genome) Sequencing Panel

ORDER OPTIONS
View Ordering Instructions1) Select Test Type
2) Select Additional Test Options
No Additional Test Options are available for this test.

